Molecular identification and nutritional analysis of green tide algae in the coastal waters of Qinhuangdao, China
- 1Key Laboratory of Marine Eco-Environmental Science and Technology, First Institute of Oceanography, Ministry of Natural Resources, Qingdao, China
- 2Laboratory of Marine Ecology and Environmental Science, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao, China
- 3Marine Ecological Restoration and Smart Ocean Engineering Research Center of Hebei Province, Qinhuangdao, China
- 4North China Sea Ecological Center of the Ministry of Natural Resources, Qingdao, China
Since 2015, the coastal waters of Qinhuangdao in Hebei province, China have experienced green tide disasters for consecutive years. Resource utilization is an important measure for preventing and controlling green tides. To comprehensively utilize green tide algae, this study first identified theses green tide algae and then assessed their nutritional value. Based on analysis of the internal transcribed spacer (ITS), 5S spacer regions (5S) and the mitochondrial cytochrome C oxidase I (COI), the dominant species were Ulva prolifera, Bryopsis plumose, Ulva australis, and Gracilariopsis lemaneiformis. The basic nutritional components, i.e., chlorophyll, amino acids, mineral elements, and heavy metal contents, of these four algae were measured and analyzed, and the four dominant species were shown to be high-nutrient-value marine foods with complete and reasonable protein, amino acid, and mineral compositions. This indicates that there is value in further research and development towards their utilization as nutrient sources. This study provides a basis for the comprehensive development and utilization of green tide algae resources in the coastal waters of Qinhuangdao.
1 Introduction
Green tides are a type of harmful algal bloom that commonly occur in coastal areas globally (Liu et al., 2013; Zhou et al., 2015). Large-scale green tides have occurred in China (Wang et al., 2015), the Philippines (Largo et al., 2004), and Japan (Yabe et al., 2009). In recent years, the distribution and frequency of green tides have increased worldwide (Xiao et al., 2021), becoming a global marine environmental problem (Yu et al., 2023). Since 2015, there have been frequent green tides in the coastal waters of Qinhuangdao, China, with blooms occurring from the middle of April to late September (Han et al., 2022a). Unlike the green tide in the Yellow Sea, the origin and primary occurrence area of the green tide in Qinhuangdao is the Jinmenghaiwan bathing beach and its adjacent waters (Song et al., 2019b; Han et al., 2022a). The attached macroalgae on seaweed beds in the waters adjacent to this beach are the main source of the green tide (Song et al., 2019a). The accumulation and decay of a large biomass of algae in the coastal waters of these beaches have seriously hindered tourist activities, negatively impacting the tourism industry of Qinhuangdao and posing a huge threat to the marine environment of the Beidaihe Scenic Area (Song et al., 2019a; Han et al., 2022a). Unlike the single dominant species in the green tide of the Yellow Sea, in Qinhuangdao the free-floating macroalgae are a mixture of various green and red algae species (Song et al., 2019a; Han et al., 2022b).
With the development of molecular biology techniques, an increasing number of researchers are using these methods for algae identification to make up for the deficiencies of traditional classification methods. DNA barcoding technology is a powerful tool for algal identification (Hebert and Gregory, 2005; Saunders, 2005; McDevit and Saunders, 2009). Currently, the DNA barcoding methods applied to macroalgae include the following genes: rbcL (a large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase from the chloroplast genome) (Saunders and McDevit, 2012), COI (cytochrome c oxidase subunit I), UPA (a fragment of the 23S rRNA gene, a universal plasmid amplification region) (Saunders and Moore, 2013), LSU (28S ribosomal RNA large subunit sequence), ITS (internal transcribed spacer) (Hayden et al., 2003), and tufA (elongation factor subunit in chloroplast genome). Wang et al. (2018) investigated and analyzed the species composition of green tide algae in the southern Yellow Sea using ITS and 5S sequences and examined the trends in early floating green algae succession from 2009 to 2017. Zhao et al. (2013) studied 31 red algae samples from the Qingdao intertidal zone using the COI, UPA, and ITS genes and found that COI and UPA genes could be used to distinguish red algae samples.
Green tides have caused serious losses to coastal aquaculture and tourism (Huo et al., 2013; Liu et al., 2016; Zhang et al., 2019). Additionally, the stench emitted from the decay of green tide algae on the beach has caused serious social problems. It is necessary to collect green tide algae promptly and dispose of or treat the biomass. Analyzing the nutritional composition of green tide algae and evaluating the safety of their use as a resource are important to allow for resource utilization treatment of green tide disasters. There are economically valuable algae in green tide algae that are nutritious and can be used for food or medicine. By processing them accordingly, waste can be turned into a valuable resource and a detriment can be converted into a benefit. In this study, ITS, 5S and COI sequences were used to identify green tide algae in the coastal waters of Qinhuangdao. The basic nutritional composition, amino acids, mineral elements, and heavy metal elements were measured, providing an empirical basis for comprehensively developing and utilizing green tide algae resources in a targeted manner.
2 Materials and methods
2.1 Sample collection
According to the occurrence of green tide in Qinhuangdao, fresh green tide algae samples were collected in June 2020 from the coastal waters of Jinmenghaiwan bathing beach. The samples were stored at low temperature (4-6°C) and transported to the laboratory within 48h. Some of the collection of 15 samples of green algae (Gr) on 28th of June 2020 and 15 samples of red algae (Rd) on 29th of June 2020 were used for the morphological and molecular identification of macroalgae. The remaining samples were rinsed multiple times with seawater to remove sediment and other debris and then rinsed with fresh water to remove salt. Algal thalli with a uniform morphology were selected and dried at 55°C for 48 h, ground into a powder with a grinder, sieved, and sealed in sample bags that were stored in a refrigerator at 4°C for later analysis of their nutritional composition.
2.2 Morphological identification of macroalgae
The macroalgae were rinsed three times with sterile seawater to remove sand, sediment, debris, and epiphytes from their surfaces. They were then cultured for three days before morphological identification of species based on color, shape, branching, and cell arrangement.
2.3 Molecular identification of macroalgae
Total genomic DNA was extracted according to the instructions of the Plant Genome DNA Kit (Tiangen Biotech, Beijing, China). The ITS and 5S sequences were used as gene markers for green algae samples, and the COI gene sequence was selected as the gene marker for red algae samples. The PCR primers used for the three gene markers are shown in Table 1. The 20 μL PCR reaction consisted of 1 uL of template DNA, 12 μL of ddH2O, 2 μL of 10× PCR buffer, 1.6 μL of MgCl2, 1.2 μL of dNTPs, 0.2 μL of Taq polymerase, and 1 μL each of forward and reverse primers. The PCR products of the seven genes were detected using 3% agarose gel electrophoresis. The purified DNA was sequenced bidirectionally at Shanghai Sangon Biotech Co., Ltd. The PCR thermocycling protocols for ITS, 5S, and COI were performed as described by Song et al. (2019b).
2.4 Phylogenetic analysis
The similarity of the target sequence was determined by BLAST on the NCBI server (http://www.ncbi.nlm.nih.gov/BLAST/). Phylogenetic trees were constructed using the maximum likelihood method implemented in Mega v.5.10 (Tamura et al., 2007).
25 Methods of determining nutritional components
All nutritional components were quantified according to the Chinese Standard from the National food safety standard for food analysis (GB). Crude protein was determined by Kjeldahl method (GB 5009.5-2016). 0.2 g of the treated sample were transfered to digestion tubes, added catalyst and 10 mL of concentrated sulfuric acid on the digestion furnace, and digested it at 420°C. and then measured it with a FOSSKjeltec2300 Kjeldahl nitrogen analyzer after cooling. Crude fat was determined by Soxhlet extraction method (GB 5009.6-2016). Ash content was determined by muffle furnace ashing method (GB 5009.4-2016). The treated sample were burned at 550°C. The crude fiber was determined by gravimetric method (GB/T 5009.10-2003). The moisture content was determined by direct drying method (GB 5009.3-2016), an appropriate amount of sample was weighed in a constant weight evaporating dish and baked in an oven at 105°C for 1 h, dried to a constant weight. Carbohydrates were determined based on the difference from these contents, calculated as 100 - (moisture + protein + fat + ash).
Chlorophyll and carotenoids were determined by acetone extraction. The chlorophyll a, chlorophyll b and carotenoid contents were calculated according to Eqs. (1), (3) and (4), and the final results were expressed in mg/g.
The amino acid composition of the protein was determined by hydrolyzing the samples with 6 mol/L hydrochloric acid; 17 amino acids were then determined using a Hitachi L-8900 fully automatic amino acid analyzer (Hitachi, Tokyo, Japan) (GB 5009.124-2016). Amino acids were analyzed under the following conditions: the injection volume was 20 μL, the flow rate of pump 1 was 0.4 mL/min at 10.5 MPa, the flow rate of pump 2 was 0.35 mL/min at 0.8 MPa, the separation column temperature was 50°C, and the reaction column temperature was 136°C.
The inorganic elements lead, Zn (Zinc), Ca (Calcium), Fe (Iron), Cu (Copper), P (Phosphorus), Mn (Manganese), Pb (Lead), Cd (Cadmium), Cr (Chromium) and Hg (Mercury) in green tide algae, was determined by copper specific method reference reference GB 5009.268-2016, taked the sample in the digestion tank, added 5 mL of nitric acid, digested it at 420°C., fixed volume to 50 mL, and analyzed by the instrument inductively coupled plasma emission spectrometer (Thermo Fisher ICAP7200 Duo).
3 Results
3.1 Morphological identification of macroalgae
Ulva australis (formerly Ulva pertusa) (Chlorophyta) thallus coloration was slightly yellow, and the edge of the thallus had wrinkles. There were irregular small holes on the surface of the thallus, and the edge was relatively thin (Figure 1A). For Bryopsis plumosa (Chlorophyta), the thallus was dark green. The thallus had obvious main branches and irregular side branches that were similar to feathers (Figure 1B). The thallus of Ulva prolifera was fresh green in color and tubular, with many branches; the base was often composed of fine filaments (Figure 1C). Gracilariopsis lemaneiformis (Rhodophyta) had a purple-red, cylindrical thallus with many branches (Figure 1D). Arranged closely, the cells were rectangular or polygonal in shape. There were one to three starch granules in each cell (Figure 1E). The cells were not densely arranged; cells were conical in shape (Figure 1F). The cells were arranged regularly and were polygonal with rounded corners. There was one starch granule in each cell (Figure 1G). The cells were irregularly arranged with gaps between them, and the cells varied greatly in size and shape (Figure 1H).
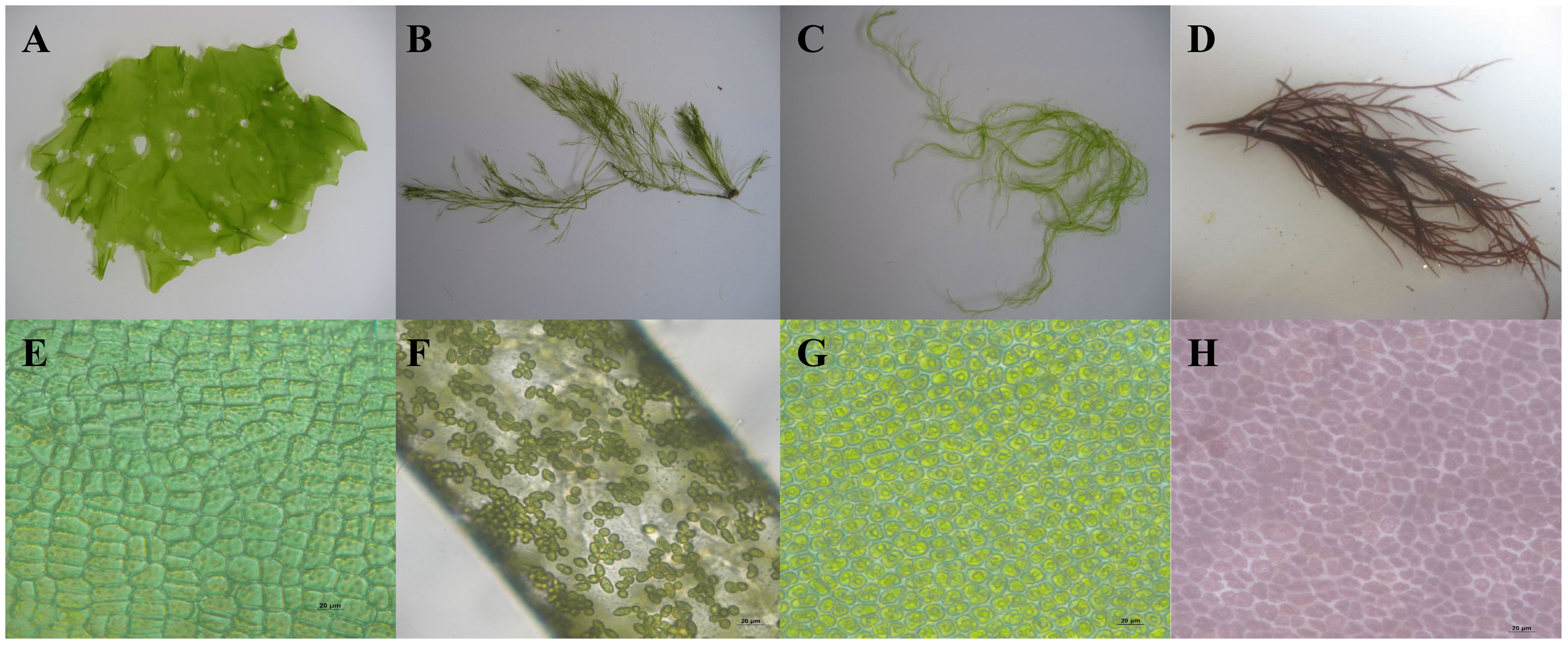
Figure 1 Morphological characteristics of the macroalgal (A and E: Ulva australis; B and F, Bryopsis plumosa; C and G: Ulva prolifera; D and H: Gracilariopsis lemaneiformis) (Zeng et al., 2023).
3.2 Molecular identification
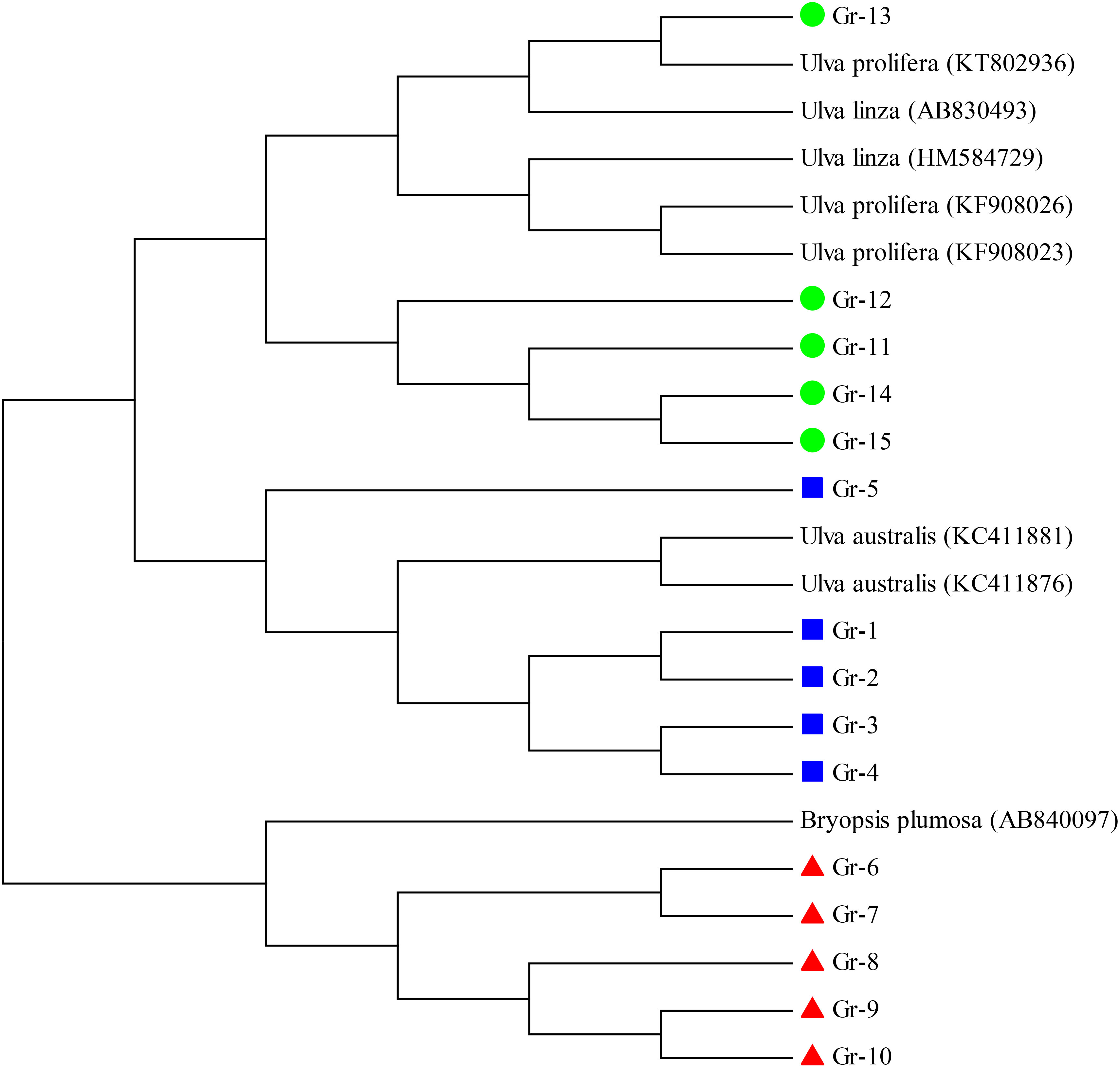
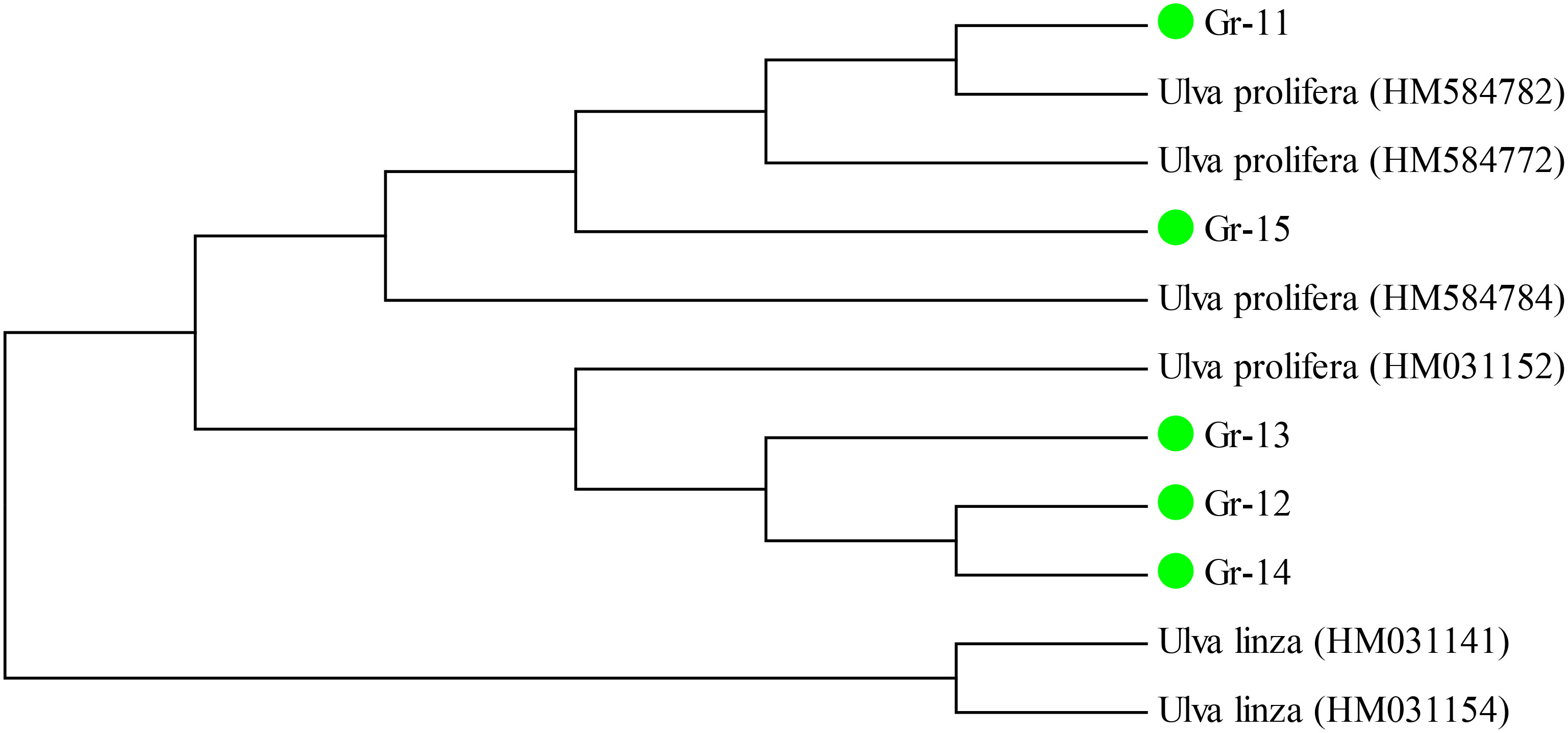
The green algae samples were classified into three different branches (Figure 2). Samples Gr1-Gr5 clustered as U. australis, while samples Gr6-Gr10 had the highest similarity to B. plumosa (JF894104) and clustered within the same branch. Samples Gr11-Gr15 clustered within the branch of U. prolifera/linza. The phylogenetic tree based on 5S sequences, as shown in Figure 3, clearly divided the U. prolifera/linza branch into two groups and all samples clustered within the U. prolifera branch. The phylogenetic tree based on COI sequences showed that all 15 red algal samples collected during the green tide outbreak clustered within the G. lemaneiformis branch (Figure 4). According to the phylogenetic tree based on 5S, ITS, and COI sequences, the dominant algal species in green tide in Qinhuangdao were identified as U. australis, U. prolifera, B. plumosa, and G. lemaneiformis.
In a comprehensive analysis of the molecular identification results and the morphological characteristics of the macroalgae, U. australis, B. plumosa, U. prolifera, and G. lemaneiformis were confirmed to be the dominant green tide species. The ITS, 5S and COI genes can serve as the appropriate gene markers for molecular identification of green tide algae in the coastal waters of Qinhuangdao.
3.3 Basic nutritional components in the dominant green tide algal species
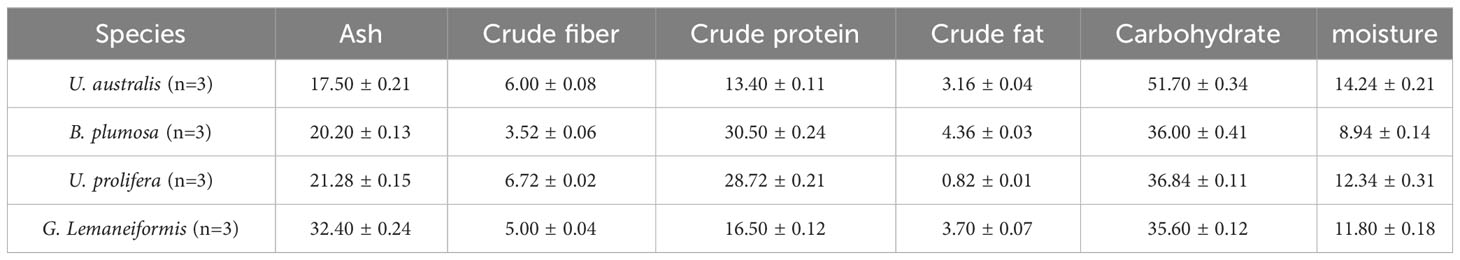
The general nutrient content of algae refers to the crude protein, fat, carbohydrates, ash and other substances in the dried seaweed. The four green tide algae had high crude protein content, with B. plumosa having the highest (30.5%) and U. australis having the lowest (13.4%) (Table 2). The crude fat content in the four green tide algae was relatively low, accounting for less than 5% of their dry weight. The four green tide algae had different ash content, with G. Lemaneiformis having the highest (32.4%) and U. australis having the lowest (17.5%). Carbohydrates were the main nutritional components in these algae, accounting for 35.6% to 51.7% of their dry weight. The four green tide algae also had relatively high crude fiber content, accounting for about 5% of their dry weight. Modern medicine and nutrition have determined that crude fiber has many important physiological impacts, so much so that it is considered as the “seventh nutrient” in addition to the traditional six essential nutrients, which has been confirmed by a large number of factual studies and epidemiological investigations both at home and abroad (Xie et al., 2009). Overall, the four green tide algae found in the coastal waters of Qinhuangdao appear to be a promising algae resource owing to their high protein, low fat, and high fiber content.
3.4 Chlorophyll and carotenoid content in the dominant green tide algal species
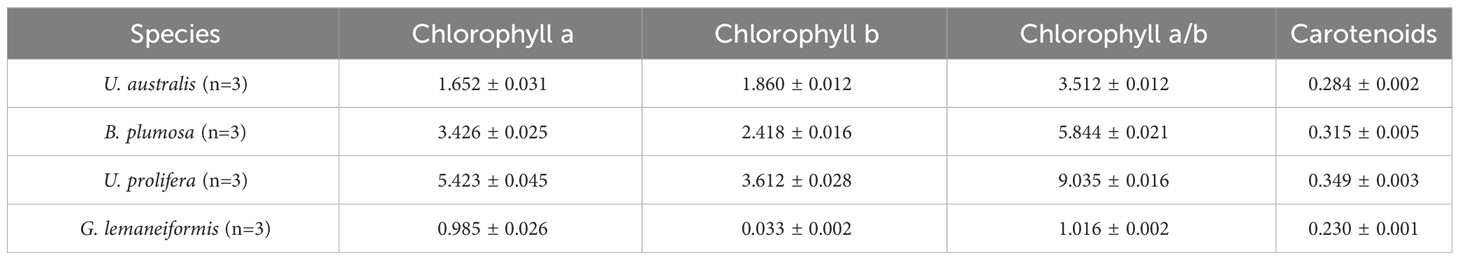
As shown in Table 3, the content of chlorophyll a and b was higher in samples of U. prolifera and B. plumosa and lower in samples of G. lemaneiformis and U. australis. The content of carotenoids was similar across the four dominant species (~0.3 mg/g). The total chlorophyll content differed greatly between the four species, with U. prolifera having the highest total chlorophyll content (9.035 mg/g) and G. lemaneiformis having the lowest (1.016 mg/g).
3.5 Amino acid content in the dominant green tide algal species
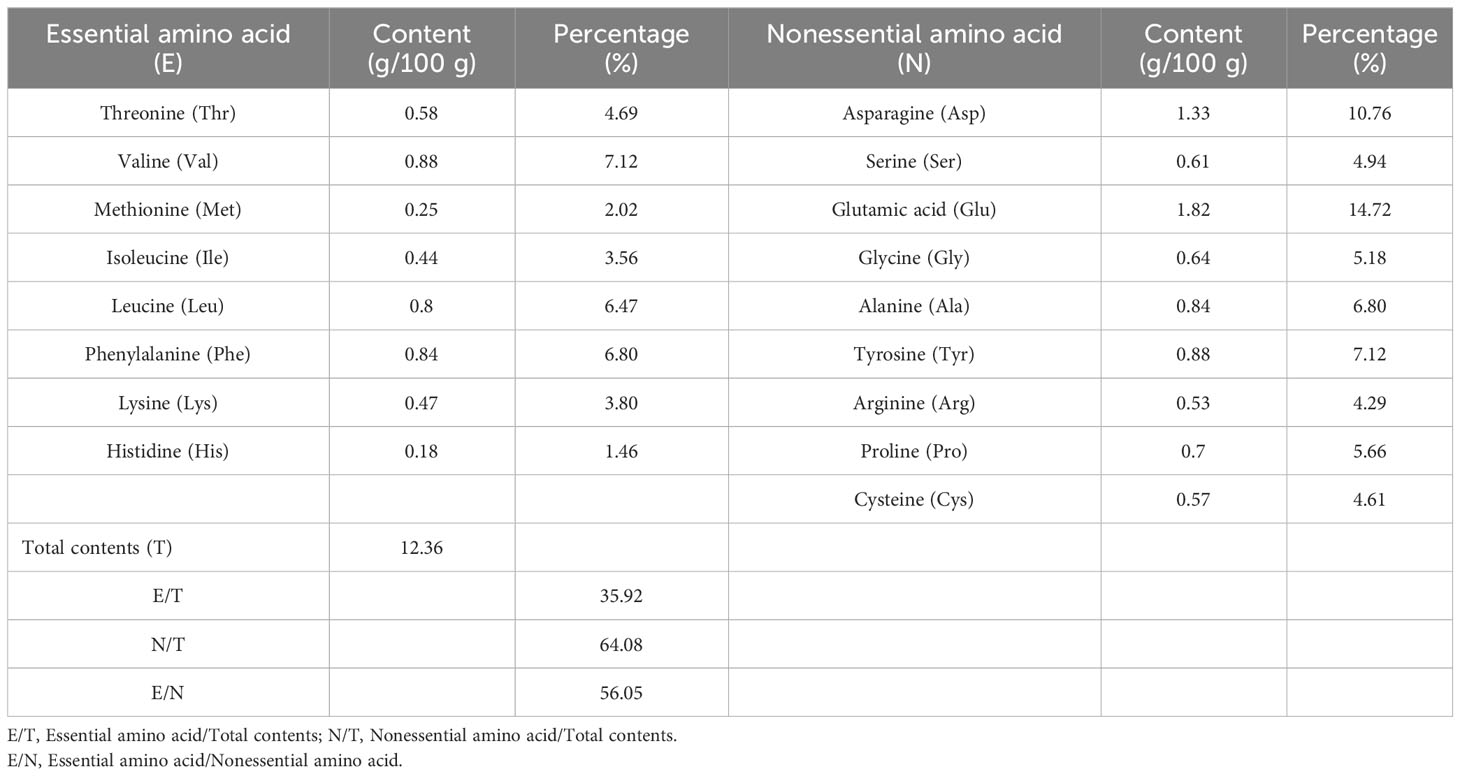
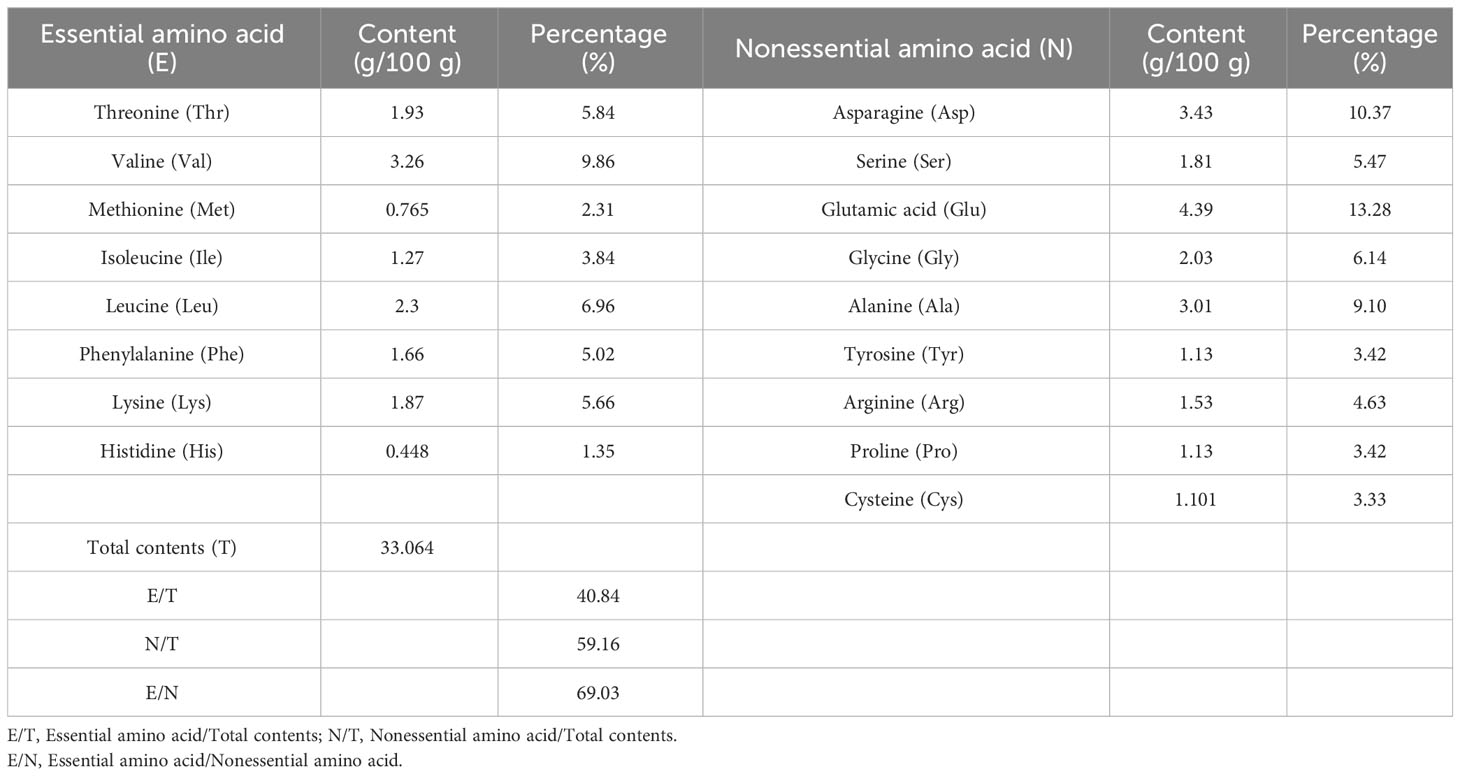
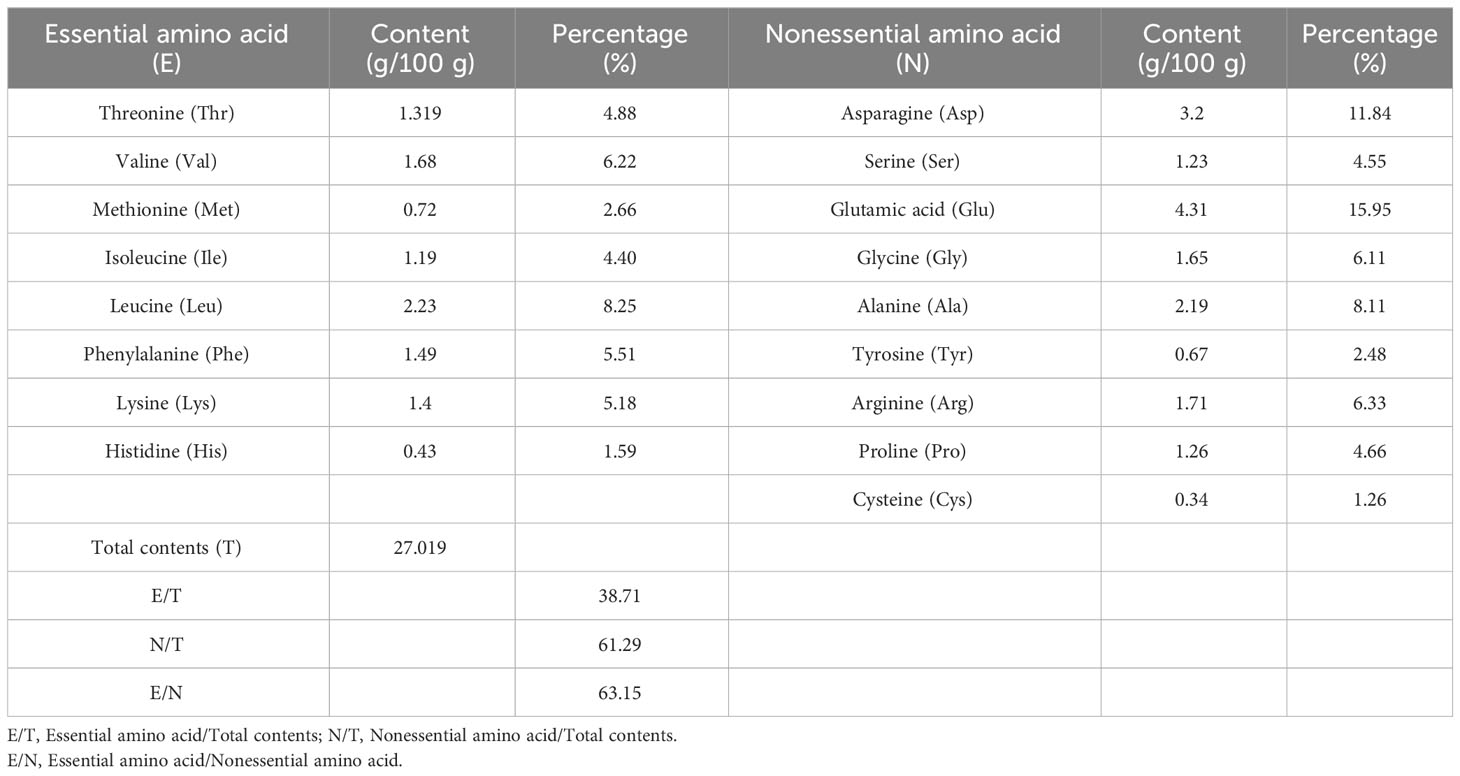
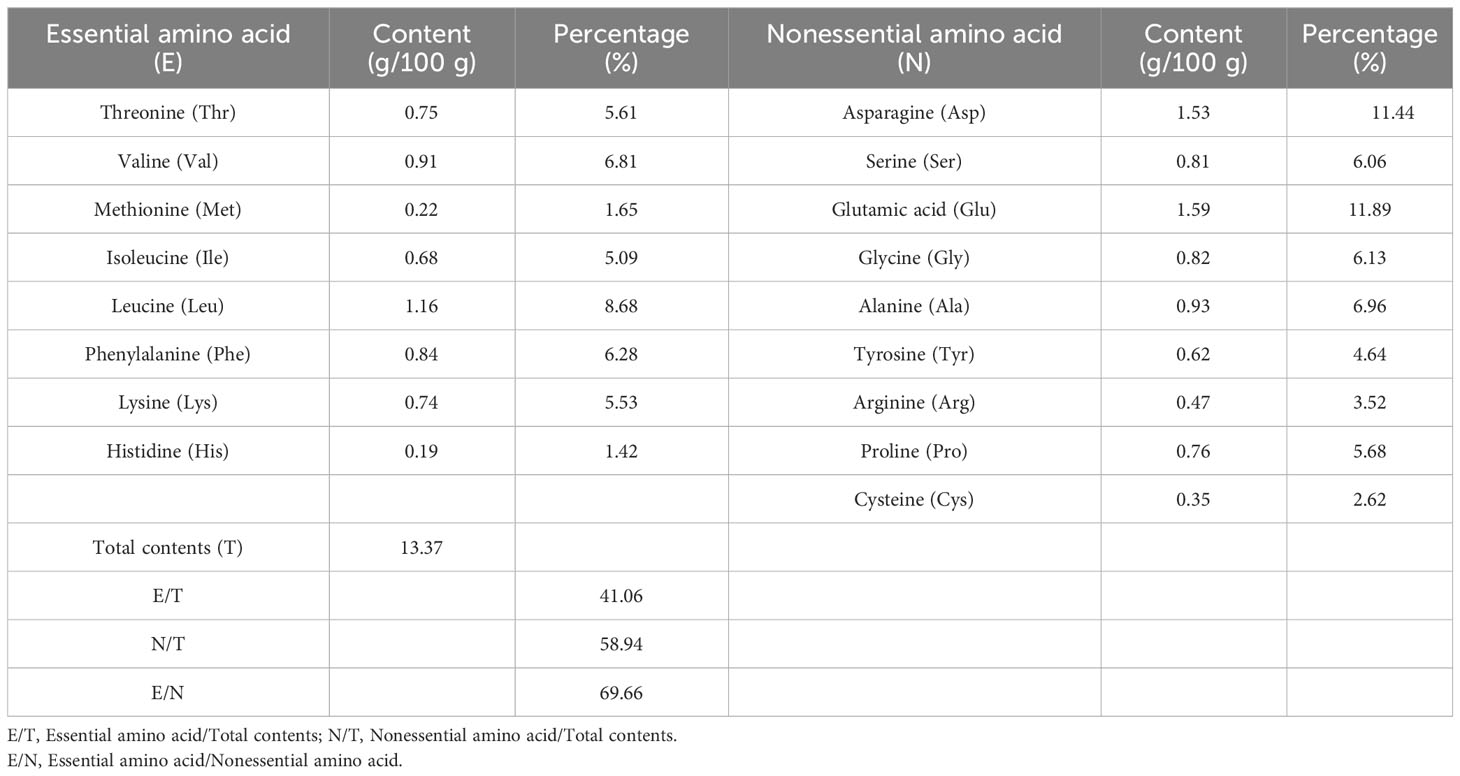
The quality of protein mainly depends on the type and content of amino acids, and whether the ratio of essential amino acids conforms to the pattern of human dietary protein is an important indicator for evaluating the quality of protein. As shown in Tables 4–7, the ratios of the total amounts of the eight essential amino acids to the total amounts of amino acids (EAA/TAA) in U. prolifera, G. lemaneiformis, U. australis, and B. plumosa were 38.71%, 41.06%, 35.92%, and 40.84%, respectively. The ratios of essential amino acids to non-essential amino acids (EAA/NEAA) were 63.15%, 69.66%, 56.05%, and 69.03%, respectively. These ratios are close to the ideal amino acid composition pattern recommended by the FAQ/WTO (EAA/TAA ≥ 40%, EAA/NEAA ≥ 60%). The average EAA/TAA ratios of G. lemaneiformis and B. plumosa were both higher than 40%, higher than those seen in U. prolifera and U. australis. In addition, the content of umami amino acids (glutamic acid, asparagine, glycine, and alanine) in the four species of green tide algae was higher than 40%. The content of umami amino acids accounted for 46.67% of the total content of amino acids in U. prolifera, which is highest than that in G. lemaneiformis (42.1%), U. australis (43.12%), and B. plumosa (42.31%). Among these amino acids, the contents of glutamic and aspartic acid were the highest. The abundance of umami amino acids is one of the reasons why U. prolifera has a rich flavor. Green tide algae should therefore have a strong umami flavor and could be used as a natural seasoning for food and animal feed. The protein quality of green tide algae was good, with a high amino acid content, complete amino acid types, and high proportions of essential amino acids, and they were rich in umami amino acids. This indicates that they could be excellent resources worth further research and development.
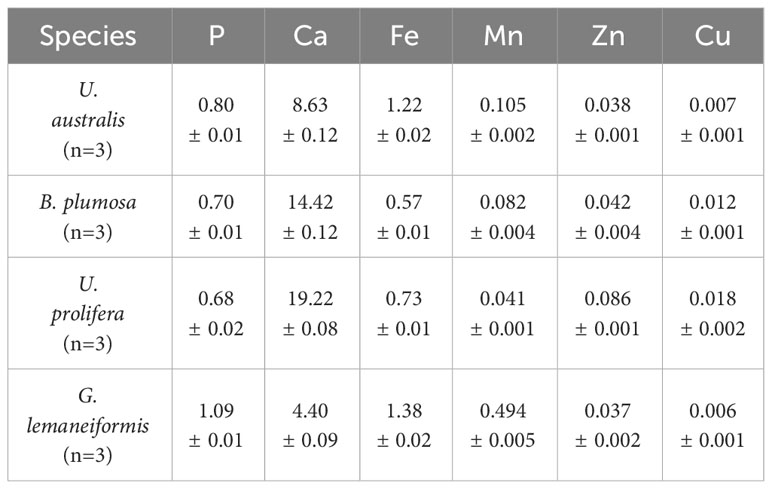
3.6 Analysis of P, Ca, Zn, Fe, Cu, and Mn in the dominant green tide algal species
The four macroalgae contained a large amount of Ca and were rich in P, Fe, and the other elements assessed, including Zn and Cu (Table 8). Fe is the main component of hemoglobin as well as many different enzymes and plays an important role in tissue respiration and biological oxidation. Cu participates in the hematopoietic process and can prevent anemia, while Zn is involved in the synthesis of many enzymes, accelerates growth and development, and promotes intellectual development. The high content of mineral elements found in green tide algae appears to meet the mineral needs of the human body, indicating their potential for development and utilization as food or medicine.
3.7 Analysis of heavy metals (Pb, Cd, Cr, and Hg) in the dominant green tide algal species
Algae accumulate marine metal ions in their bodies through active transport and passive absorption, and it is particularly important to analyze the heavy metal content of marine algae and evaluate the safety. Table 9 shows the content of heavy metal elements found in the green tide algae. No heavy metal exceeded the limits listed in “The National Standard for Algae and Its Products” (GB19643-2016), indicating that the green tide algae meet safety standards for consumption, which could be used for food processing and other resource utilization.
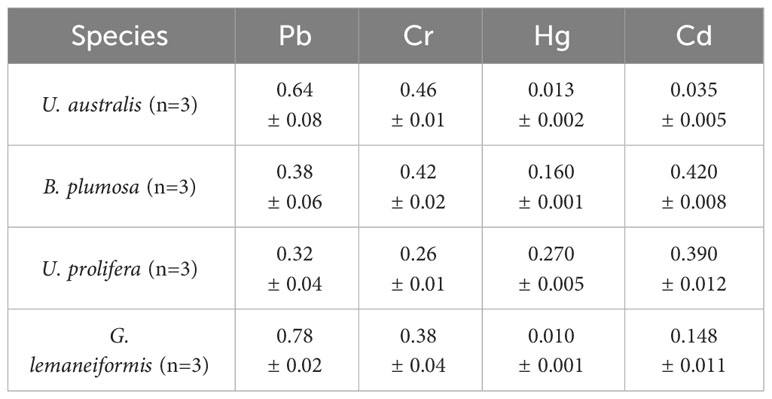
Table 9 The heavy metal element content of the four species of green tide algae (mg/kg) (Mean ± SD).
4 Discussion
4.1 Dominant species of green tide algae in the coastal waters of Qinhuangdao
In the past few decades, green tide outbreaks have expanded globally (Zhou et al., 2015). Since 2015, green tides have erupted in the coastal waters of Qinhuangdao every year (c; Han et al., 2022a). Accurately identifying the dominant species of algae present is the first step in understanding this marine ecological disaster. However, It is difficult to accurately identify algae based on morphological characteristics alone (Wang et al., 2010). Thus, in this study, morphological assessment and molecular identification have been used in combination to identify the dominant species of green tide algae in the coastal waters of Qinhuangdao.
According to the phylogenetic analysis based on the ITS sequences, samples Gr1-Gr5 belonged to the U.australis, which also is a species that causes green tides in East Asia frequently (Xiao et al., 2013). Samples Gr5-Gr10 clustered with B. plumose, which was also supported by morphological characteristics. B. plumosa is a common alga in the intertidal zone of the northern Yellow and Bohai Seas (Shao et al., 2011). For the dominant species of green tide in the coastal waters of Qinhuangdao, the ITS gene separated U. australis from B. plumosa. The samples Gr11-Gr15 were clustered in the U. prolifera/linza branch according to the phylogenetic analysis based on the ITS sequences. Previous studies have shown that ITS sequences cannot effectively determine the taxonomic status of these species in the LPP branch (Shen et al., 2012; Xiao et al., 2013). A more informative DNA marker, the 5S rDNA sequence, has been reported to be more effective in further distinguishing these species (Shimada et al., 2008). Based on the phylogenetic tree constructed using the obtained 5S rDNA sequences, all the green tide algae samples from Qinhuangdao were identified to be U. prolifera, which was supported by the morphological characteristics of these samples. The phylogenetic tree constructed based on COI sequences showed that the red algae samples were clustered in the G. lemaneiformis branch.
4.2 Analysis of the nutritional composition of green tide algae in Qinhuangdao
Since its first occurrence, green tides in Qinhuangdao have received much attention from the local government. During the green tide bloom, the Qinhuangdao municipal government expends a large amount of manpower and material resources to clean up the algae accumulated on the shores. The cleared algae are generally piled at the garbage treatment station on the west side of Jinmenghaiwan bathing beach where they are prone to rot, causing secondary pollution. According to reports from local authorities, the annual specialized labor costs for algae clearing alone reach nearly 5 million RMB (Song et al., 2019a). Researchers widely agree that removing algae from the source or utilizing the large biomass of accumulated algae as a resource are two effective methods for disaster prevention and reduction of the green tide consequences (Liu et al., 2013). Since the large-scale occurrence of green tide in the Yellow Sea, disaster prevention and reduction have mainly focused on reducing green algae entering the sea and collecting floating green algae with specialized collection equipment (Liu et al., 2013). The collected algae are mainly processed into bio-organic fertilizer through composite enzymatic hydrolysis and algal chelation (Wang et al., 2014).
Because the formation mechanism of the green tide in Qinhuangdao has not yet been fully clarified, effective source control measures cannot be implemented (Song et al., 2019b). In addition, the algae in the coastal waters of Qinhuangdao are mainly suspended at depths 0–20 m away from the shoreline, where the shallow water and suspension characteristics make it difficult to effectively use collection equipment (Han et al., 2019; Song et al., 2019a). Green tide algae have certain economic value and can be utilized according to their different characteristics. Currently, processing these algae into seaweed fertilizer is a common resource utilization method (Wang et al., 2014). In the present study, a preliminary quantitative analysis of the basic nutritional components of green tide algae in Qinhuangdao was conducted, aiming to lay an empirical foundation for the development and utilization of green tide algae as resources. The four dominant green tide species in the coastal waters of Qinhuangdao were inferred to have a strong algal flavor and could be used as natural seasoning for human food and animal feed. According to the ideal FAO/WHO dietary model, the amino acid composition of high-quality protein requires a ratio of essential amino acids to total amino acids between 40% and 60%, and a ratio of essential amino acids to non-essential amino acids above 60%. For green tide algae in Qinhuangdao, the two ratios are close to these requirements, indicating that they have good-quality protein. Not only do these algae have a high amino acid content and a complete variety of amino acids, but they also have a high proportion of essential amino acids and are rich in umami amino acids, making them a good source of plant protein. In addition, aspartic acid in green tide algae plays an important role in the energy and nitrogen metabolism of mitochondria, the production of central nervous system excitatory neurotransmitters, and the urea cycle. It is widely used in the clinical treatment of hepatitis, cirrhosis, and hepatic coma. The high content of aspartic acid in green tide algae identified here would likely have important health benefits (Cai et al., 2009). Algae is rich in nutrients, in addition to protein and other components, but also contains essential sodium, potassium and other trace elements (Yao et al., 2016). Potassium and sodium work together to maintain the water balance in the body and the normal function of neuromuscular. Potassium can eliminate harmful substances in the body to a certain extent through the kidneys, which is an indispensable substance in the human body.
In this study, The basic nutritional components, i.e., chlorophyll, amino acids, mineral elements, and heavy metal contents, of these four algae were measured and analyzed. The results of this study also shows that in the green tide algae in Qinhuangdao, the heavy metal content of Pb, Cd, Cr, and Hg did not exceed safety limits specified in international standards “Algae and algae products-Food and feed applications-General overview of limits, procedures and analytical methods” (PD CEN/TR 17559:2021). This all indicates that these green tide algae found meet food safety standards and could be developed and utilized. There is no lack of nutritious economic algae with edible and medicinal values in green tide algae, which can be processed into food and medicine and utilized, so as to achieve the purpose of turning waste into treasure and turning harm into benefit.
5 Conclusions
Green tides can cause various environmental problems, and at the same time, the odors emanating from the decay of these algae also cause serious social problems (Han et al., 2019; Song et al., 2019a). Therefore, it is necessary to salvage the green tide algae in time and treat them harmlessly, and the analysis of the nutrient composition of green tide algae and the evaluation of the safety of resource utilization is an important aspect of the harmless treatment of the green tide disaster. In this study, U. australis, B. plumosa, U. prolifera, and G. lemaneiformis were the dominant species of the green tide in the coastal waters of Qinhuangdao. These four dominant species were identified to be highly nutritious marine foods with complete compositions of proteins, amino acids, and mineral elements. These green tide algae in the coastal waters of Qinhuangdao are thus promising species meriting further research and development. At present, the scale of resource utilization of green tide algae in China is relatively small, and the economic benefits of the industry are poor. State and local governments should guide and encourage enterprises to strengthen the development of green tide algae resource utilization and provide policy support and preferential treatment for these ventures, especially in sectors related to food, feed, and medicine. Through this approach, achieving sustainable development of this waste product while protecting the marine ecological environment can be realized.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YL: Writing – original draft, Supervision, Resources, Funding acquisition, Data curation. XM: Writing – original draft, Supervision, Resources, Methodology. MJ: Writing – original draft, Supervision, Resources, Methodology. WS: Writing – original draft, Methodology, Data curation. GW: Writing – original draft, Methodology, Data curation. HH: Writing – original draft, Supervision, Data curation, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Marine Ecological Restoration and Smart Ocean Engineering Research Center of Hebei Province (HBMESO2319); The Open Research Fund of Shandong Provincial Key Laboratory of Marine Ecological Environment and Disaster Prevention and Mitigation (202205); National Key Research and Development Program of China under contract No. (2019YFC1407902).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Cai C. E., Yao B., Shen W. R., He P. M. (2009). Determination and analysis of nutrition compositions in Enteromorpha clathrata. J. Shanghai Ocean Univ. 18 (2), 155–159. (in Chinese with English abstract). doi: 10.1360/972009-754
GB/T 5009.10-2003 (2003). determination of crude fiber in vegetable foods [S]. (Beijing: Standards Press of China).
GB 19643-2016 (2016). national food safety standard algae and its products [S]. (Beijing: Standards Press of China).
GB 5009.124-2016 (2016). National food safety standard determination of amino acids in food [S]. (Beijing: Standards Press of China).
GB 5009.268-2016 (2016). national food safety standard determination of multi-elements in food [S]. (Beijing: Standards Press of China).
GB 5009.3-2016 (2016). national food safety standard Determination of moisture in food [S]. (Beijing: Standards Press of China).
GB 5009.4-2016 (2016). National food safety standard determination of ash in food [S]. (Beijing: Standards Press of China).
GB 5009.5-2016 (2016). national food safety standard determination of protein in food [S]. (Beijing: Standards Press of China).
GB 5009.6-2016 (2016). national food safety standard determination of fat in food [S]. (Beijing: Standards Press of China).
Han H. B., Li Y., Ma X. J., Song W., Wang Z. L., Zhang X. L. (2022a). Factors influencing the spatial and temporal distributions of green algae micro-propagules in the coastal waters of Jinmenghaiwan, Qinhuangdao, China. Mar. pollut. Bull. 175, 113328. doi: 10.1016/j.marpolbul.2022.113328
Han H. B., Li Y., Ma X. J., Song W., Wang Z. L., Zhang X. L., et al. (2022b). Population differentiation in the dominant species (Ulva prolifera) of green tide in coastal waters of China. Acta Oceanol. Sin. 41 (11), 108–114. doi: 10.1007/s13131-022-1985-5
Han H. B., Song W., Wang Z. L., Ding D. W., Yuan C., Zhang X. L., et al. (2019). Distribution of green algae micro-propagules and their function in the formation of the green tide in the coast of Qinhuangdao, the Bohai Sea, China. Acta Oceanol. Sin. 38 (08), 72–77. doi: 10.1007/s13131-018-1278-1
Hayden H. S., Blomster J., Maggs C. A., Silva P. C., Stanhope M. J., Waaland J. R., et al. (2003). Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur. J. Phycol. 38 (3), 277–294. doi: 10.1080/1364253031000136321
Hebert P. D., Gregory T. R. (2005). The promise of DNA barcoding for taxonomy. Syst. Biol. 54 (5), 852–859. doi: 10.1080/10635150500354886
Huo Y. Z., Zhang J. H., Chen L. P., Hu M., Yu K. F., Chen Q. F., et al. (2013). Green algae blooms caused by Ulva prolifera in the southern Yellow Sea: Identification of the original bloom location and evaluation of biological processes occurring during the early northward floating period. Limnol. Oceanogr. 58 (6), 2206–2218. doi: 10.4319/lo.2013.58.6.2206
Largo D. B., Sembrano J., Hiraoka M., Ohno M. (2004). Taxonomic and ecological profile of green tides species of Ulva (Ulvales, Chlorophyta) in central Philippines. Hydrobiologia 5 (12), 247–253. doi: 10.1023/B:HYDR.0000020333.33039.4b
Liu D. Y., Keesing J., He P. M., Wang Z. L., Shi Y. J., Yang Y. J. (2013). The world's largest macroalgae bloom in the Yellow Sea, China: Formation and implications. Estuar. Coast. Shelf Sci. 129, 2–10. doi: 10.1016/j.ecss.2013.05.021
Liu X. Q., Wang Z. L., Zhang X. L. (2016). A review of the green tides in the Yellow Sea, China. Mar. Environ. Res. 119, 189–196. doi: 10.1016/j.marenvres.2016.06.004
McDevit D. C., Saunders G. W. (2009). On the utility of DNA barcoding for species differentiation among brown macroalgae (Phaeophyceae) including a novel extraction protocol. Phycol. Res. 57 (2), 131–141. doi: 10.1111/j.1440-1835.2009.00530.x
Saunders G. W. (2005). Applying DNA barcoding to red macroalgae: a preliminary appraisal holds promise for future appli-cations. Philos. T.R. Soc B. 360 (1462), 1879–1888. doi: 10.1098/rstb.2005.1719
Saunders G., Kucera H. (2010). An evaluation of rbcL, tufA, UPA, LSU and its as DNA barcode markers from the marine green macroalgae. Cryptogamie Algol. 31, 487–528.
Saunders G. W., McDevit D. C. (2012). Methods for DNA barcoding photosynthetic protists emphasizing the macroalgae and diatoms. Methods Mol. Biol. 858, 207–222. doi: 10.1007/978-1-61779-591-6_10
Saunders G. W., Moore T. E. (2013). Refinements for the amplification and sequencing of red algal DNA barcode and RedTol phylogenetic markers: a summary of current primers, profiles and strategies. Algae 28 (1), 31–43. doi: 10.4490/algae.2013.28.1.031
Shao K. S., Gong N., Li K., Wang Z. L., Li D. C. (2011). Nitrogen and phosphorus nutrition physiology of Enteromorpha linza and Bryopsis plumosa (Cholorophyta). Haiyang Xuebao 33, 131–139. (in Chinese with English abstract). doi: 10.1088/1674-1137/35/2/019
Shen Q., Li H., Li Y., Wang Z. L., Liu J. S., Yang W. D. (2012). Molecular identification of green algae from the rafts based infrastructure of Porphyra yezoensis. Mar. pollut. Bull. 64 (10), 2077–2082. doi: 10.1016/j.marpolbul.2012.07.021
Sherwood A. R., Chan Y. L., Presting G. G. (2010). Application of universally amplifying plastid primers to environmental sampling of a stream periphyton community. Mol. Ecol. Resour. 8 (5), 1011–1014. doi: 10.1111/j.1755-0998.2008.02138.x
Shimada S., Yokoyama N., Arai S., Hiraoka M. (2008). Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J. Appl. Phycol. 20 (5), 979–989. doi: 10.1007/s10811-007-9296-y
Song W., Han H. B., Wang Z. L., Li Y. (2019b). Molecular identification of the macroalgae that cause green tide in the Bohai Sea, China. Aquat. Bot. 156, 38–46. doi: 10.1016/j.aquabot.2019.04.004
Song W., Wang Z. L., Li Y., Han H. B., Zhang X. L. (2019a). Tracking the original source of the green tide in the Bohai Sea, China. Estuar. Coast. Shelf Sci. 219, 354–362. doi: 10.1016/j.ecss.2019.02.036
Tamura K., Dudley J., Nei M., Kumar M. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Wang S. Y., Huo Y. Z., Zhang J. H., Cui J. J., Wang Y., Yang L. L., et al. (2018). Variations of dominant free-floating Ulva species in the source area for the world’s largest macroalgal blooms, China: Differences of ecological tolerance. Harmful Algae 74, 58–66. doi: 10.1016/j.hal.2018.03.007
Wang J. F., Jiang P., Cui Y., Li N., Wang M. Q., Lin H. Z., et al. (2010). Molecular analysis of green-tide-forming macroalgae in the Yellow Sea. Aquat. Bot. 93 (1), 25–31. doi: 10.1016/j.aquabot.2010.03.001
Wang Z. L., Xiao J., Fan S. L., Liu X. Q., Liu D. Y. (2015). Who made the world’s largest green tide in China?—an integrated study on the initiation and early development of the green tide in Yellow Sea. Limnol. Oceanogr. 60, 1105–1117. doi: 10.1002/lno.10083
Wang J., Zhan Q. Y., Zheng S., Zheng X. L., Yang Q. Y., Wang P. (2014). Preparation of Bio-Fertilizer from Enteromorpha prolifera and its effect on the quality of brassica campestris ssp. chinensis. J. Ocean U. China 44 (1), 62–67. (in Chinese with English abstract). doi: 10.16441/j.cnki.hdxb.2014.01.010
Xiao J., Li Y., Song W., Wang Z. L., Fu M. Z., Li R. X., et al. (2013). Discrimination of the common macroalgae (Ulva and Blidingia) in coastal waters of Yellow Sea, northern China, based on restriction fragment-length polymorphism (RFLP) analysis. Harmful Algae 27, 130–137. doi: 10.1016/j.hal.2013.05.003
Xiao J., Wang Z. L., Liu D. Y., Fu M. Z., Yan T. (2021). Harmful macroalgal blooms (HMBs) in China’s coastal water: Green and golden tides. Harmful Algae 107, 102061. doi: 10.1016/j.hal.2021.102061
Xie C. L., Huang J., Sun B. (2009). Chemical composition of Porphyra haitanensis(Rhodophyta, Bangiales) in China. Chin. J. Mar. Drugs 28 (1), 29–35. (in Chinese with English abstract). doi: 10.13400/j.cnki.cjmd.2009.01.006
Yabe T., Ishii Y., Amano Y., Koga T., Hayashi S., Nohara S., et al. (2009). Green tide formed byfree floating Ulva spp. atYatsu tidal flat, Japan. Limnology 10, 239–245. doi: 10.1007/s10201-009-0278-4
Yao H. Q., Wang F. J., Liu F. L., Liang Z. R., Wang W. J., Sun X. T., et al. (2016). Chemical analysis and nutritional assessment of new varieties of Saccharina japonica. Food Sci. 37 (12), 95–98. (in Chinese with English abstract). doi: 10.7506/spkx1002-6630-201612016
Yu Z. M., Tang Y. Z., Goble C. J. (2023). Harmful algal blooms in China: History, recent expansion, current status, and future prospects. Harmful Algae 129, 102499. doi: 10.1016/j.hal.2023.102499
Zeng B. X., Sun Y. G., Song W., Wang Z. L., Zhang X. L. (2023). Recurrence of the green tide in the Bohai Sea, China: A green tide caused by coastal reclamation projects. Journal of Sea Research 191, 102333. doi: 10.1016/j.seares.2022.102333
Zhang Y. Y., He P. M., Li H. M., Li G., Liu J. H., Jiao F. Y., et al. (2019). Ulva prolifera green tide outbreaks and their environmental impact in the Yellow Sea, China. Natl. Sci. Rev. 6, 825–838. doi: 10.1093/nsr/nwz026
Zhao X. B., Pang S. J., Shan T. F., Liu F. (2013). Applications of three DNA barcodes in assorting intertidal red macroalgal flora in Qingdao, China, Ocean University of China. J Ocean U. China 12, 1, 139–114. doi: 10.1007/s11802-013-2052-9
Keywords: Qinhuangdao, green tide algae, molecular identification, nutritional value, dominant species
Citation: Li Y, Ma X, Jiang M, Song W, Wang G and Han H (2023) Molecular identification and nutritional analysis of green tide algae in the coastal waters of Qinhuangdao, China. Front. Mar. Sci. 10:1293002. doi: 10.3389/fmars.2023.1293002
Received: 13 September 2023; Accepted: 22 November 2023;
Published: 11 December 2023.
Edited by:
Ivan Viegas, University of Coimbra, PortugalReviewed by:
Leonel Pereira, University of Coimbra, PortugalKit Leong Cheong, Guangdong Ocean University, China
Copyright © 2023 Li, Ma, Jiang, Song, Wang and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongbin Han, han10207018@163.com
 Yan Li
Yan Li Xiaojun Ma1
Xiaojun Ma1  Hongbin Han
Hongbin Han